10.8 Metabolomics Single-Omics Quick-Start Example
This section demonstrates a complete and commonly used analytical workflow for metabolomics data, from data import to functional interpretation, using example datasets.
Example data download: Github link
10.8.1 Importing Metabolomics Data
For single-omic data where no relationship table is involved, the sampleID in the abundance matrix must match those in the Sample phenotypic data.
library(EasyMultiProfiler)
meta_data <- read.table('col.txt',header = T,row.names = 1)
data <- read.table('metabol.txt',header = T,quote = '',sep = '\t')
MAE <- EMP_easy_import(data = data,coldata = meta_data,
sampleID = rownames(meta_data),
type = 'normal')
10.8.2 Exploring Metabolomics Data
View Current Metabolomics Assay
MAE |>
EMP_assay_extract() # View abundance matrix
MAE |>
EMP_coldata_extract() # View phenotype data
MAE |>
EMP_rowdata_extract() # View metabolite annotations
10.8.3 Calculate Coefficient of Variation for Quality Control (Optional)
Compute CV for Metabolite Features Using QC Samples
MAE |>
EMP_assay_extract() |>
EMP_mutate(CV = sd(c(QC1,QC2,QC3,QC4,QC5,QC6))/mean(c(QC1,QC2,QC3,QC4,QC5,QC6)),
mutate_by = 'feature',location = 'rowdata',action = 'rowwise',.before = 2)
Filter Metabolites Based on CV
MAE |>
EMP_assay_extract() |>
EMP_mutate(CV = sd(c(QC1,QC2,QC3,QC4,QC5,QC6))/mean(c(QC1,QC2,QC3,QC4,QC5,QC6)),
mutate_by = 'feature',location = 'rowdata',action = 'rowwise',.before = 2) |>
EMP_filter(feature_condition = CV < 0.3)
10.8.4 Aggregate Metabolite Data by Phenotype Annotation
Collapse Metabolites by Metabolite Name
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',
estimate_group = 'MS2Metabolite')
Collapse Metabolites by KEGG ID
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',
estimate_group = 'MS2kegg')
10.8.5 Metabolite Identifier Conversion
The EMP package supports cross-database metabolite ID conversion, including CAS, DTXSID, DTXCID, SID, CID, KEGG, ChEBI, HMDB, and Drugbank.
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',
estimate_group = 'MS2kegg') |>
EMP_feature_convert(from = 'KEGG',to = 'HMDB')
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',
estimate_group = 'MS2kegg') |>
EMP_feature_convert(from = 'KEGG',to = 'Drugbank')
10.8.6 Batch Effect Correction (Optional)
MAE |>
EMP_assay_extract() |>
EMP_adjust_abundance(.factor_unwanted = 'Region',
.factor_of_interest = 'Group',
method = 'combat_seq')
10.8.7 Metabolite Abundance Normalization
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',estimate_group = 'MS2Metabolite') |>
EMP_decostand(method = 'relative')
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',estimate_group = 'MS2Metabolite') |>
EMP_decostand(method = 'clr')
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',estimate_group = 'MS2Metabolite') |>
EMP_decostand(method = 'log2')
10.8.8 Differential Abundance Analysis
Wilcoxon Test for Differential Metabolites
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',estimate_group = 'MS2Metabolite') |>
EMP_diff_analysis(method = 'wilcox.test',estimate_group = 'Group') |>
EMP_filter(feature_condition = pvalue < 0.05)
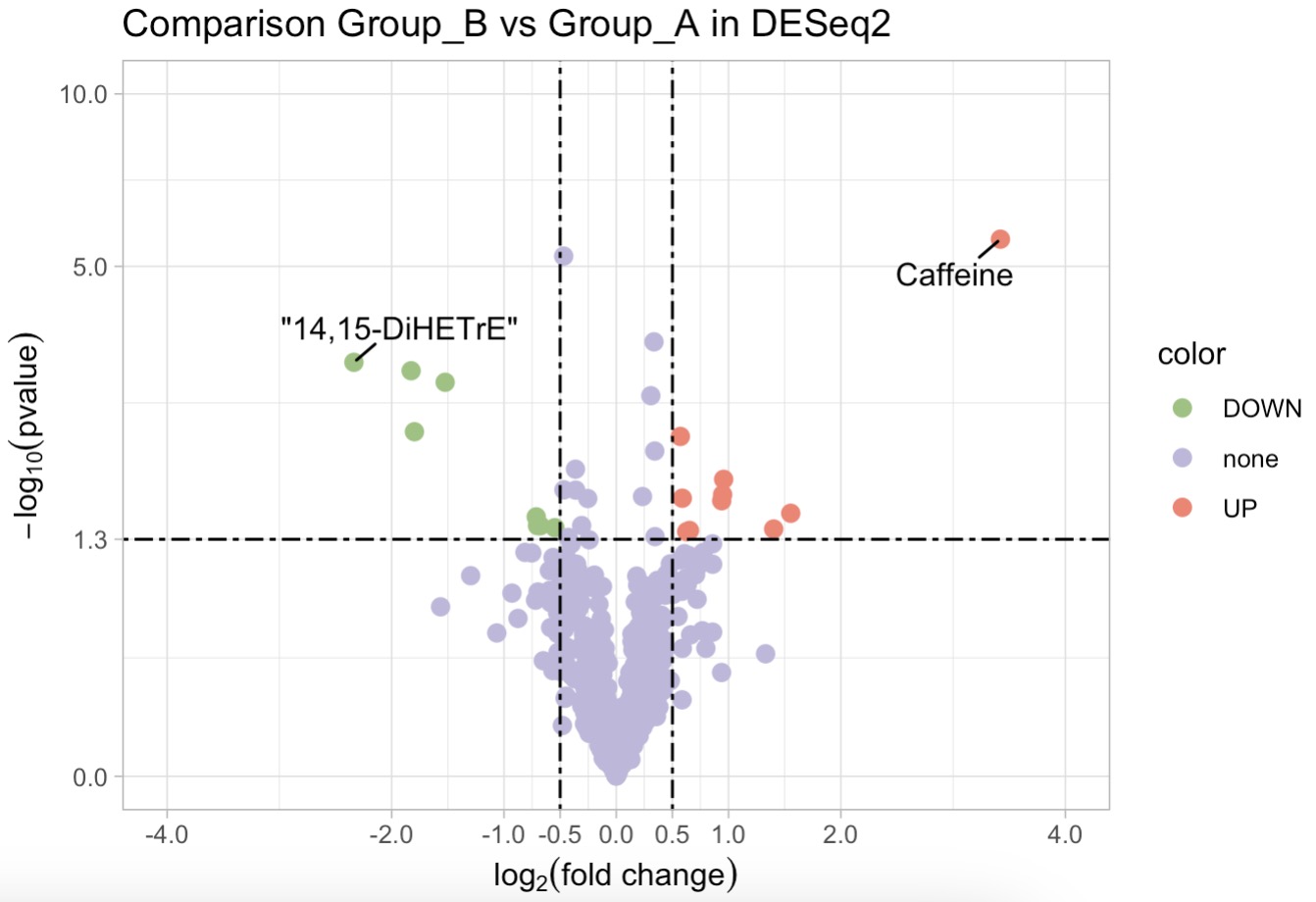
DESeq2 Analysis with Volcano Plot
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',estimate_group = 'MS2Metabolite') |>
EMP_diff_analysis(method = 'DESeq2',.formula = ~Group) |>
EMP_volcanol_plot(key_feature = c('Caffeine','\"14,15-DiHETrE\"'),
palette = c('#FA7F6F','#96C47D','#BEB8DC'),
dot_size = 2.5,threshold_x = 0.5,mytheme = "theme_light()",
min.segment.length = 0, seed = 42, box.padding = 0.5)
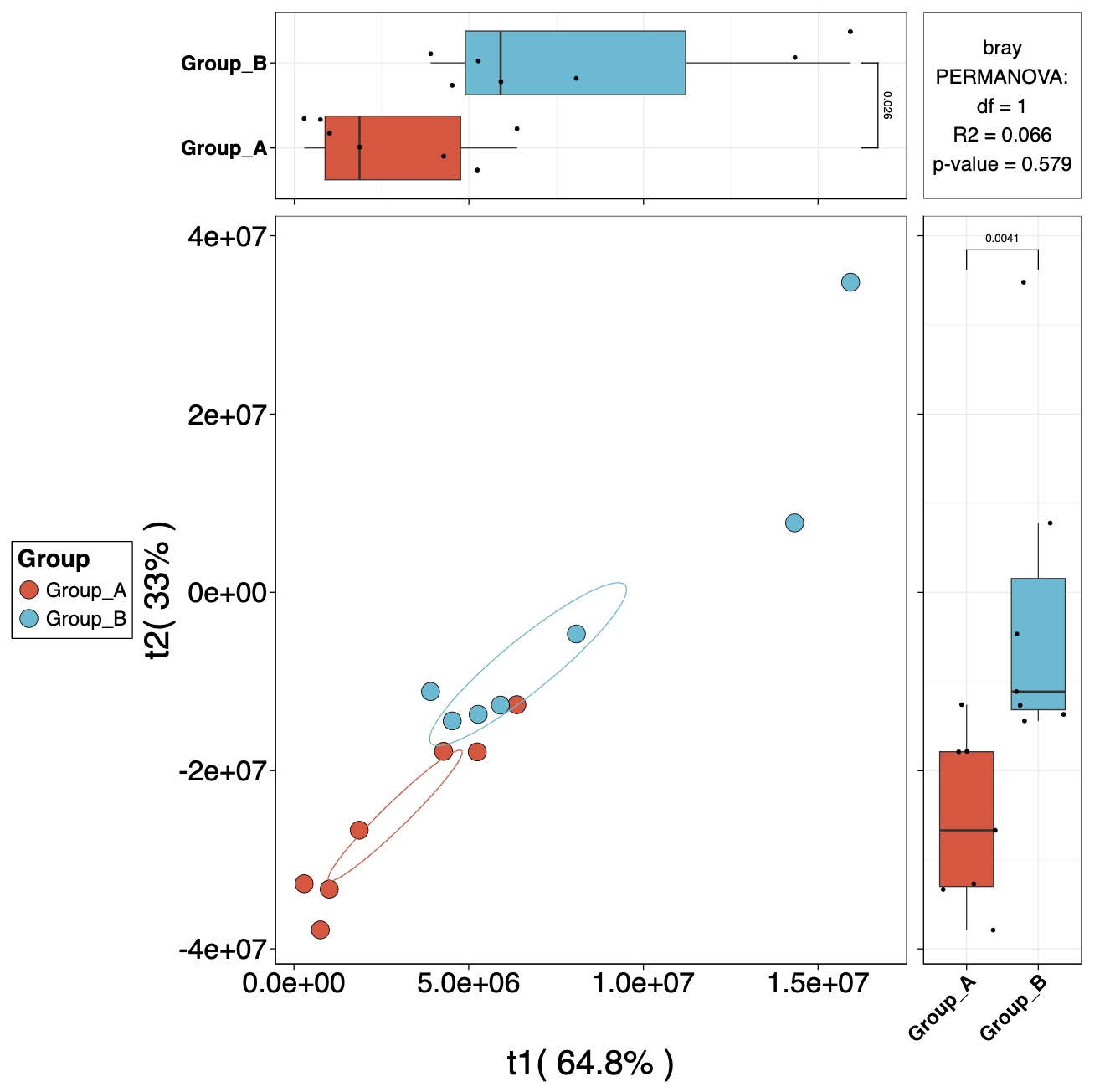
PLS and OPLS-DA Modeling
Calculate Metabolite VIP Scores
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',estimate_group = 'MS2Metabolite') |>
EMP_dimension_analysis(method = 'pls',estimate_group = 'Group')
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',estimate_group = 'MS2Metabolite') |>
EMP_dimension_analysis(method = 'opls',estimate_group = 'Group')
Visualize Dimension Reduction Models
PLS
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',estimate_group = 'MS2Metabolite') |>
EMP_dimension_analysis(method = 'pls',estimate_group = 'Group') |>
EMP_scatterplot(show='p12html',ellipse=0.3)
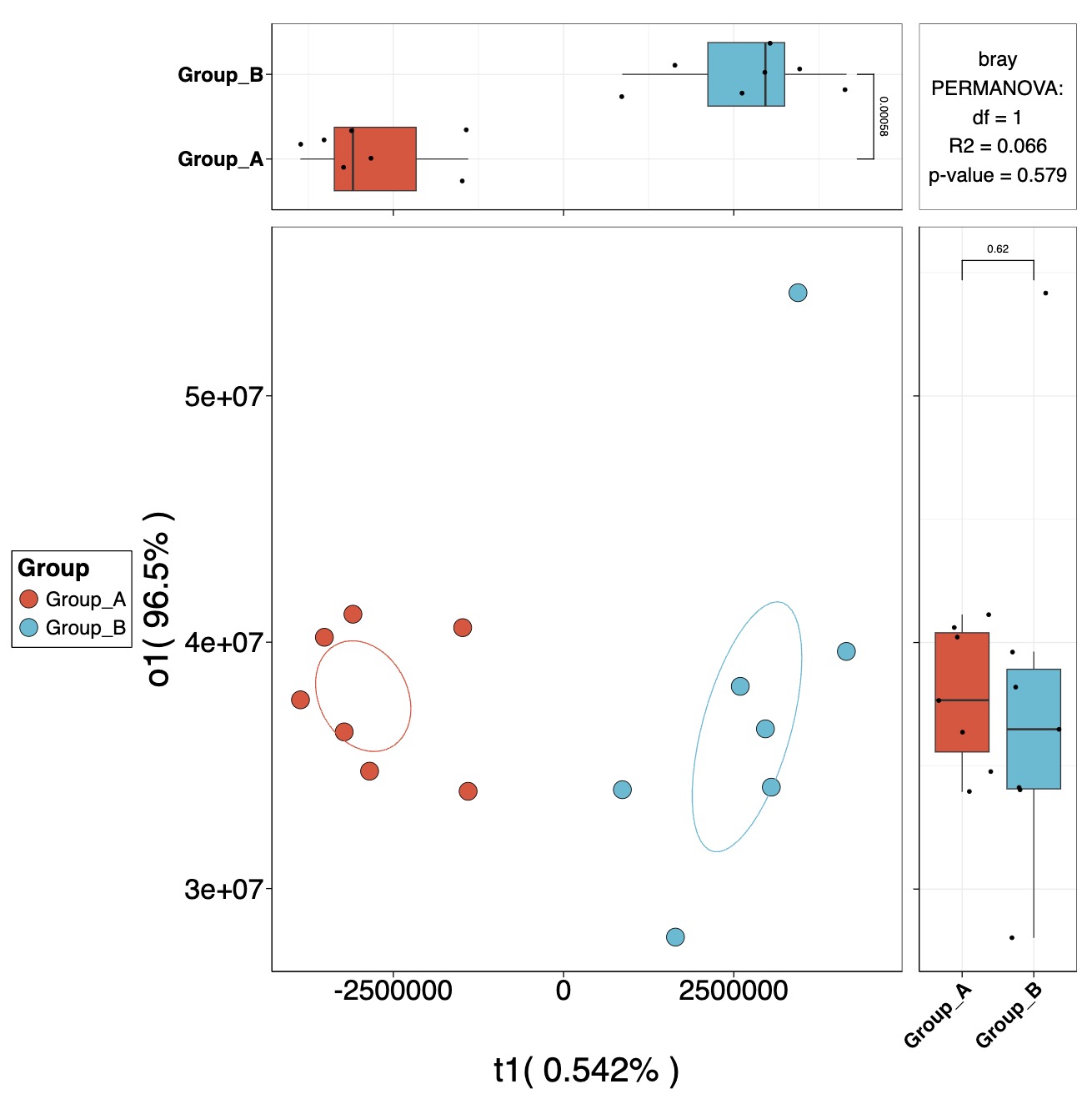
OPLS
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',estimate_group = 'MS2Metabolite') |>
EMP_dmension_analysis(method = 'opls',estimate_group = 'Group') |>
EMP_scatterplot(show='p12html',ellipse=0.3)
10.8.10 Identify Key Metabolites
Filter Metabolites Using PLS VIP and Differential Analysis This step filters features by combining results from both differential analysis and dimension reduction, with specific thresholds adjustable based on research needs.
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',estimate_group = 'MS2Metabolite') |>
EMP_diff_analysis(method = 'wilcox.test',estimate_group = 'Group') |>
EMP_dimension_analysis(method = 'pls',estimate_group = 'Group') |>
EMP_filter(feature_condition = VIP >1 & pvalue < 0.05 & abs(fold_change >1.2))
10.8.11 Machine Learning for Feature Selection
The EMP package includes Boruta, Random Forest, XGBoost, and Lasso for feature selection.
Run help(EMP_marker_analysis) for details.
Boruta Algorithm with Heatmap Visualization
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',estimate_group = 'MS2Metabolite') |>
EMP_marker_analysis(method = 'boruta',estimate_group = 'Group') |>
EMP_filter(feature_condition = Boruta_decision!= 'Rejected') |>
EMP_heatmap_plot(palette='Spectral',legend_bar='auto',
scale='standardize',
clust_row=TRUE,clust_col=TRUE)
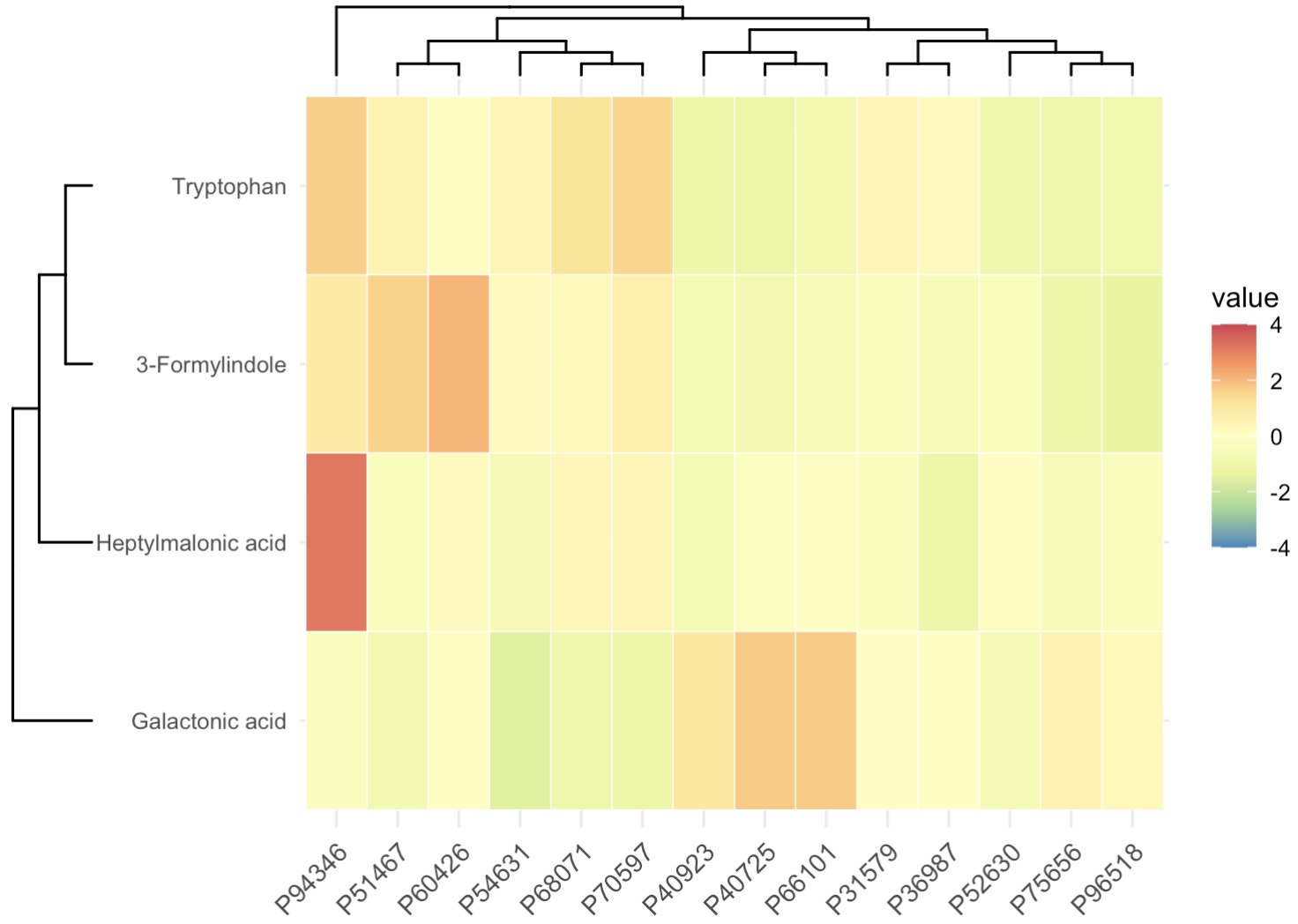
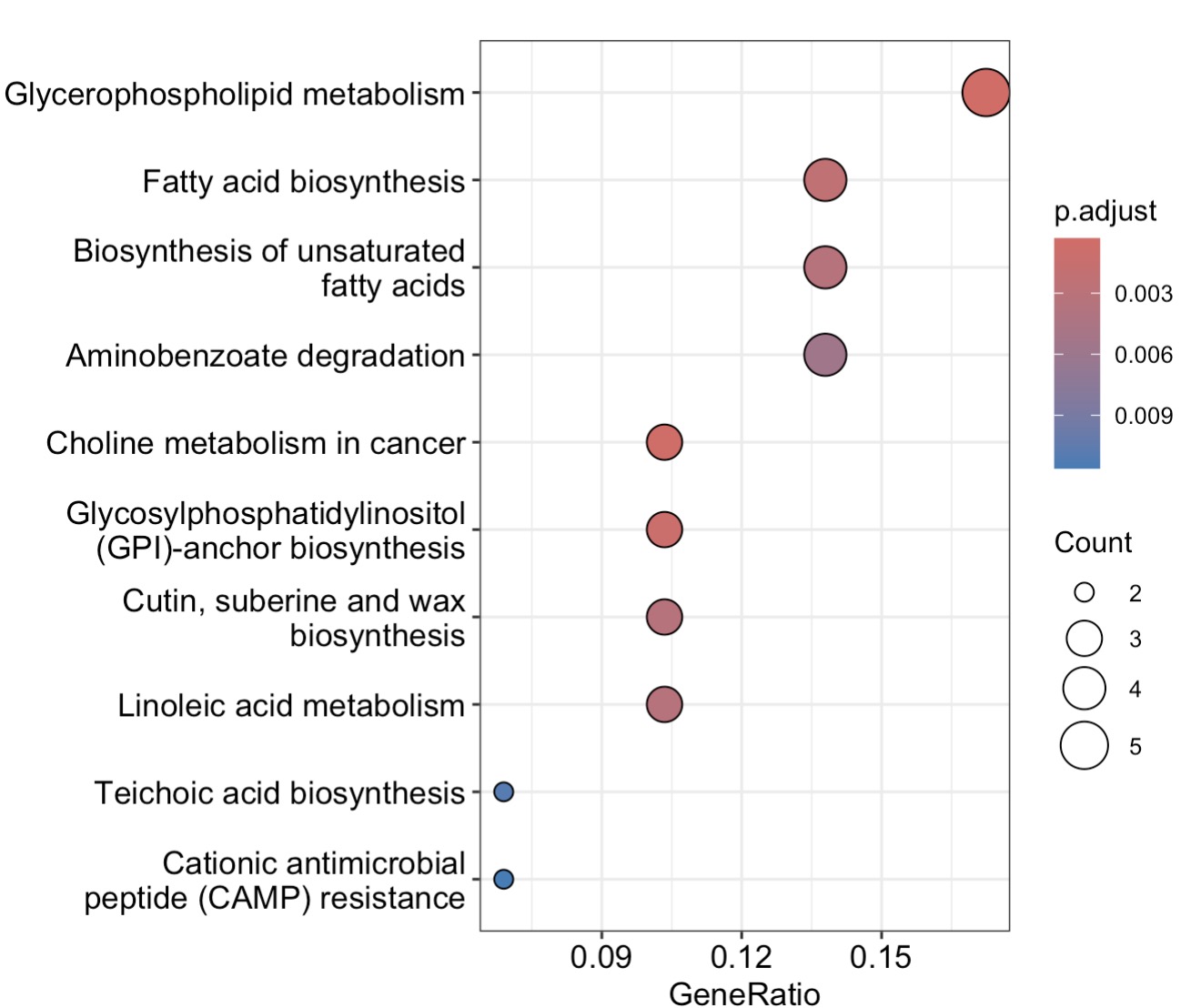
10.8.12 KEGG Enrichment Analysis for Metabolomics
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',estimate_group = 'MS2kegg') |>
EMP_dimension_analysis(method = 'pls',estimate_group = 'Group') |>
EMP_filter(feature_condition = VIP >1) |>
EMP_enrich_analysis( keyType ='cpd',
KEGG_Type = 'KEGG',
pvalueCutoff=0.05) |>
EMP_enrich_dotplot()
10.8.13 WGCNA for Metabolomics Data
Step 1: Metabolite Clustering Based on Phenotype
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',estimate_group = 'MS2kegg') |>
EMP_WGCNA_cluster_analysis(RsquaredCut = 0.8,
mergeCutHeight=0.2,
minModuleSize=10)
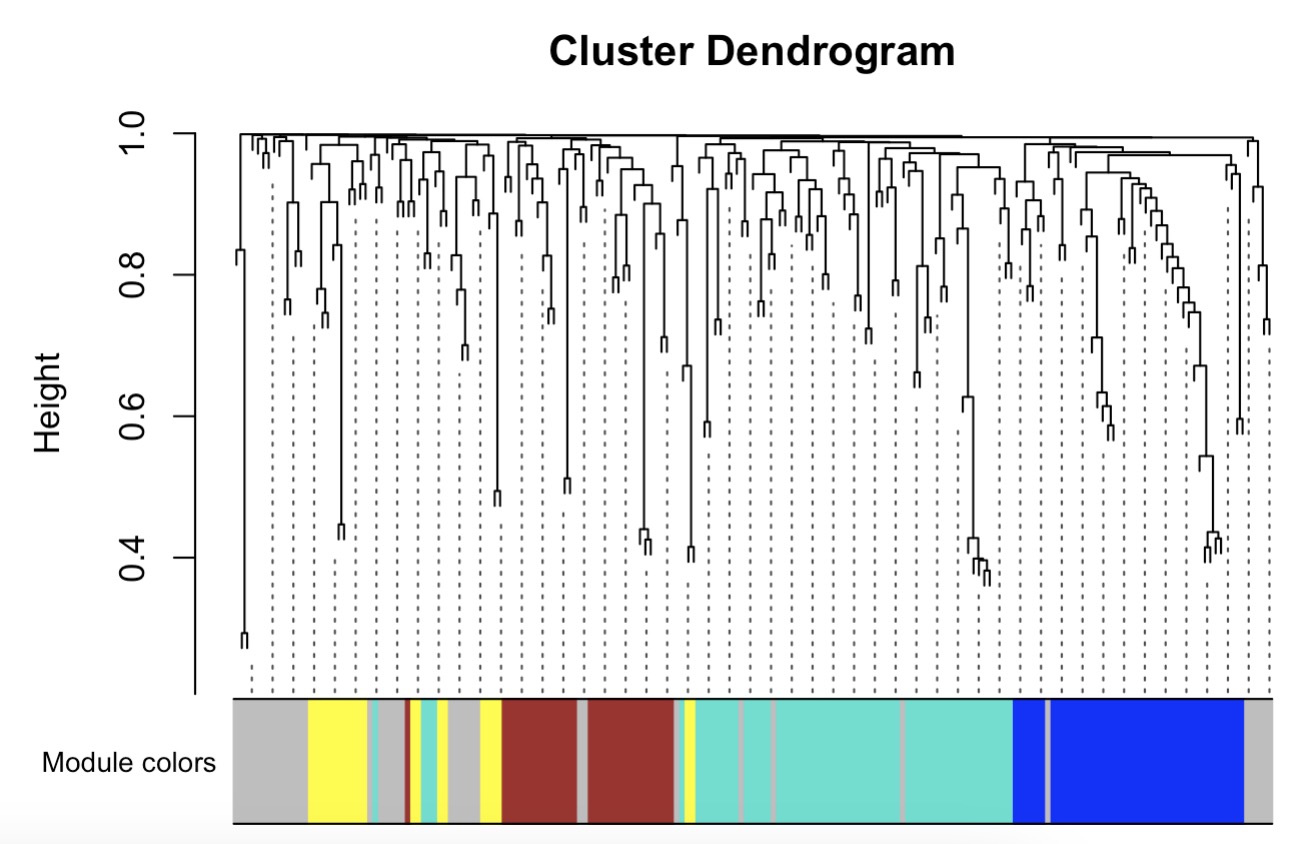
Step 2: Heatmap of Phenotype-Correlated Metabolite Modules
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',estimate_group = 'MS2kegg') |>
EMP_WGCNA_cluster_analysis(RsquaredCut = 0.8,
mergeCutHeight=0.2,
minModuleSize=10) |>
EMP_WGCNA_cor_analysis(coldata_to_assay = c('BMI','PHQ9','GAD7','HAMD','SAS','SDS'),
method='spearman') |>
EMP_heatmap_plot(palette = 'Spectral')
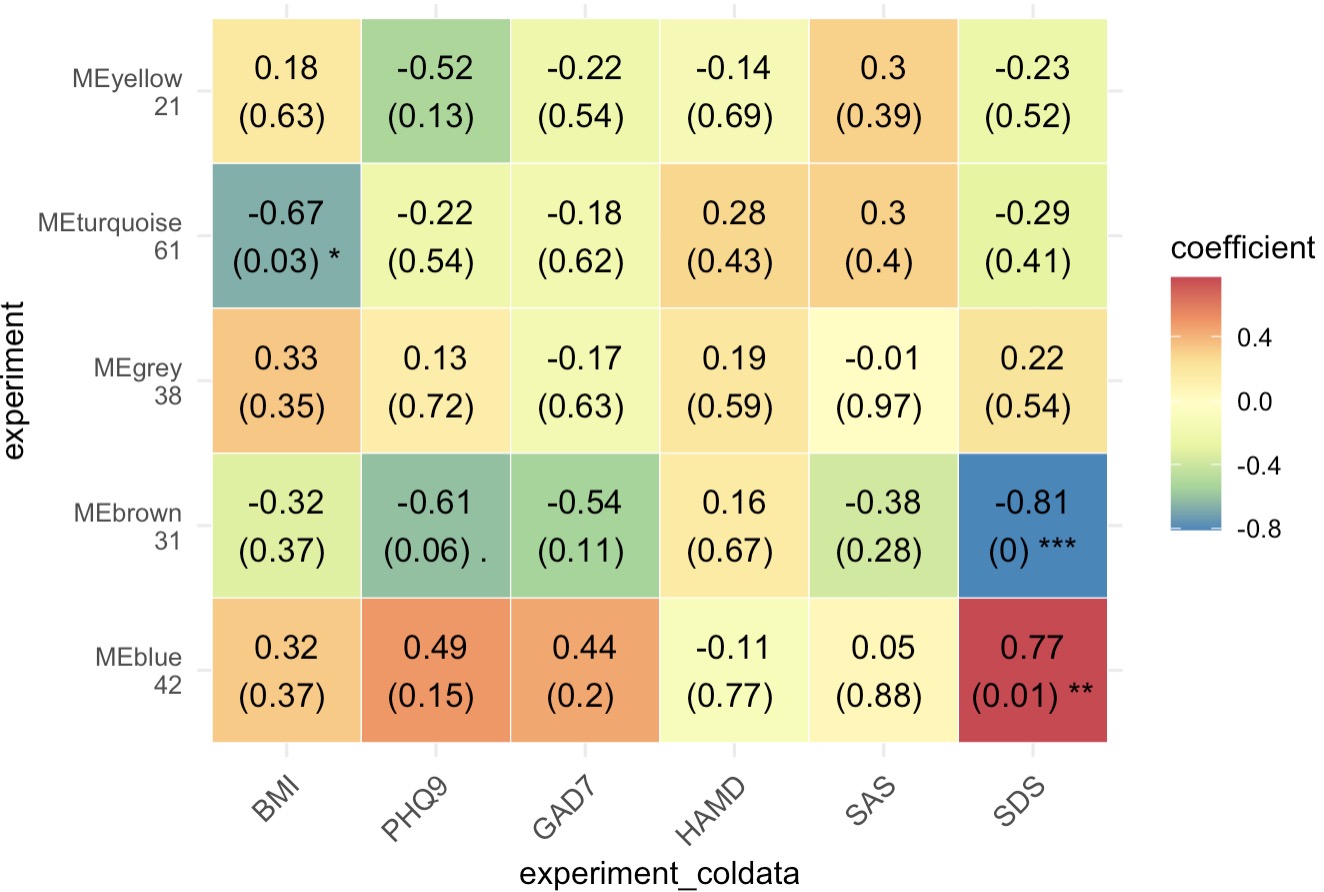
Step 3: Enrichment Analysis for Selected Metabolite Modules
MAE |>
EMP_assay_extract() |>
EMP_collapse(collapse_by = 'row',estimate_group = 'MS2kegg') |>
EMP_WGCNA_cluster_analysis(RsquaredCut = 0.8,
mergeCutHeight=0.2,
minModuleSize=10) |>
EMP_WGCNA_cor_analysis(coldata_to_assay = c('BMI','PHQ9','GAD7','HAMD','SAS','SDS'),
method='spearman') |>
EMP_heatmap_plot(palette = 'Spectral') |>
EMP_filter(feature_condition = WGCNA_color == 'blue' ) |>
EMP_enrich_analysis(keyType = 'cpd',KEGG_Type = 'KEGG') |>
EMP_enrich_dotplot()